|
The Brain Simulation Platform "Live Papers"
|
Models of Human Cortical Pyramidal Neurons
Authors: Guy Eyal, Oren Amsalem, Yoni Leibner, Matthijs B. Verhoog, Guilherme Testa-Silva, Yair Deitcher, Johannes C Lodder, Ruth Benavides-Piccione,
Juan Morales, Javier DeFelipe, Christiaan P. J. de Kock, Huibert D. Mansvelder and Idan Segev
Corresponding author: Idan Segev (
idan@lobster.ls.huji.ac.il
)
Journals:
Frontier In Cellular Neuroscience,
eLife Sciences Publications
DOI:
https://www.frontiersin.org/articles/10.3389/fncel.2018.00181/full,
https://elifesciences.org/articles/16553
Licence: the
Creative Commons Attribution (CC BY) license
applies for all files. Under this Open Access license
anyone may copy, distribute, or reuse the files as long as the authors and the original source are properly cited.
Citations:
Eyal, G., Verhoog, M. B., Testa-Silva, G., Deitcher, Y., Benavides-Piccione, R., DeFelipe, J., ...
& Segev, I. (2018). Human cortical pyramidal neurons: From spines to spikes via models. Frontiers in cellular neuroscience, 12, 181.
Eyal, G., Verhoog, M. B., Testa-Silva, G., Deitcher, Y., Lodder, J. C., Benavides-Piccione, R., ... & Segev, I. (2016). Unique membrane properties and enhanced signal processing in human neocortical neurons. Elife, 5, e16553.
Abstract:
The advanced cognitive capabilities of the human brain are often attributed to our recently evolved neocortex.
However, it is not known whether the basic building blocks of the human neocortex, the pyramidal neurons, possess unique
biophysical properties that might impact on cortical computations. Here we study this question by modelling, in details,
pyramidal cells from human neocortex, including models on their membrane properties, excitatory synapses, dendritic spines,
dendritic NMDA- and somatic/axonal Na+ spikes. This provided new insights into signal processing and computational
capabilities of these principal cells. Six human layer 2 and layer 3 pyramidal cells (HL2/L3 PCs) were modeled,
integrating detailed anatomical and physiological data from both fresh and postmortem tissues from human temporal cortex.
These cells have specific membrane capacitance (Cm) of ~0.5 µF/cm2, half of the commonly accepted 'universal' value (~1 µF/cm2)
for biological membranes; they have particularly large AMPA- and NMDA-conductances per synaptic contact
(0.88 and 1.31 nS, respectively) and a steep dependence of the NMDA-conductance on voltage. A large dataset of high-resolution
reconstructed HL2/L3 dendritic spines provided estimates for the EPSPs at the spine head (12.7 ± 4.6 mV), spine base (9.7 ± 5.0 mV),
and soma (0.3 ± 0.1 mV), and for the spine neck resistance (50–80 MΩ). Matching the shape and firing pattern of experimental
somatic Na+-spikes provided estimates for the density of the somatic/axonal excitable membrane ion channels, predicting
that 134 ± 28 simultaneously activated HL2/L3-HL2/L3 synapses are required for generating (with 50% probability) a somatic
Na+ spike. Dendritic NMDA spikes were triggered in the model when 20 ± 10 excitatory spinous synapses were simultaneously
activated on individual dendritic branches. The particularly large number of basal dendrites in HL2/L3 PCs and the distinctive cable
elongation of their terminals imply that ~25 NMDA-spikes could be generated independently and simultaneously in these cells, as
compared to ~14 in L2/3 PCs from the rat somatosensory cortex. These multi-sites non-linear signals, together with the large (~30,000)
excitatory synapses/cell, equip human L2/L3 PCs with enhanced computational capabilities. Our study provides the most comprehensive
model of any human neuron to-date demonstrating the biophysical and computational distinctiveness of human cortical neurons.
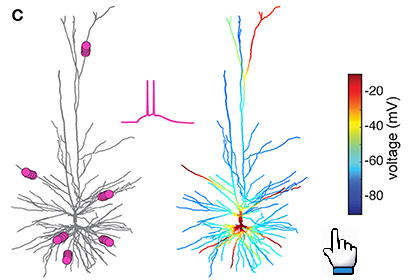
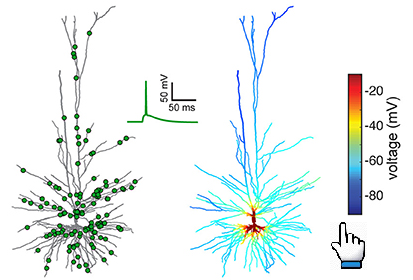
This Live Paper introduces interactively these 6 modeled human L2/3 neurons, including their morphology, experimental measurements
and modelling response to current and synaptic inputs. The models are available to download and could also be simulated on the
cloud with NEURON as a service. The user can manipulate current injections to the model and explore the voltage response at any
dendritic/somatic and axonal loci.
Model response to clusterd vs non clusterd input
Resources
Data and models: all data and models used in the paper are available at the links reported below:
-
Morphology files (in .asc format) used in the paper for each etype (see Table S5 of the Supplementary Material):
-
The
Model Catalog
of the cells